
“Unidad de Excelencia María de Maeztu”, funded by the MCIN and the AEI (DOI: 10.13039/501100011033). Ref: CEX2018-000792-M


© 2023| Department of Medicine and Life Sciences (Universitat Pompeu Fabra Barcelona) | All Rights Reserved | Powered by Scienseed
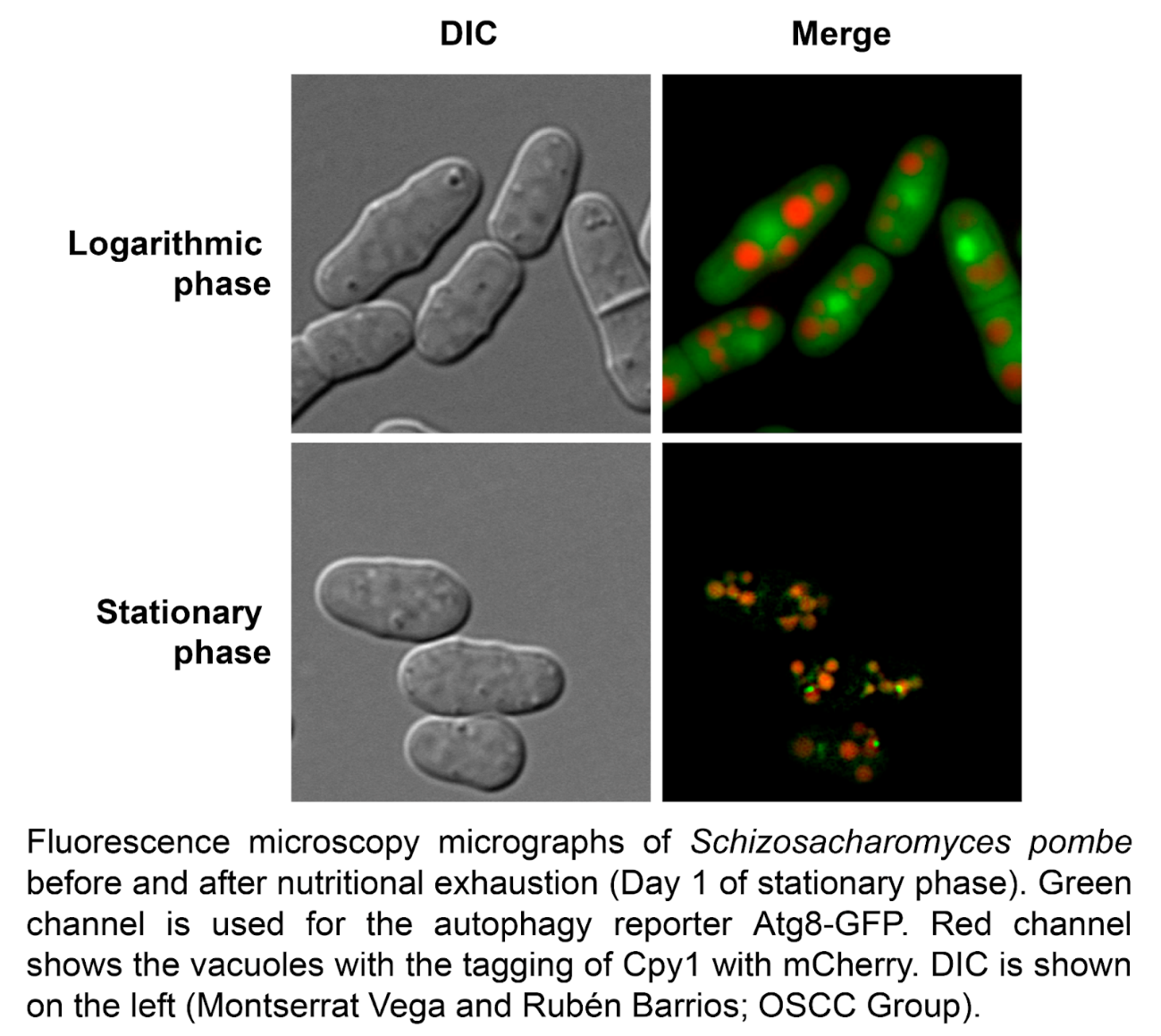
General autophagy is an evolutionarily conserved process in eukaryotes, by which intracellular materials are transported into and degraded inside lysosomes or vacuoles, with the main goal of recycling those materials during periods of starvation. The molecular bases of autophagy have been widely described in Saccharomyces cerevisiae, and the specific roles of Atg proteins in the process were first characterized in this model system. Important contributions have been made in Schizosaccharomyces pombe highlighting the evolutionary similarity and, at the same time, diversity of Atg components in autophagy. However, little is known regarding signals, pathways and role of autophagy in this distant yeast. Here, we undertake a global approach to investigate the signals, the pathways and the consequences of autophagy activation. We demonstrate that not only nitrogen but several nutritional deprivations including lack of carbon, sulfur, phosphorus or leucine sources, trigger autophagy, and that the TORC1, TORC2 and MAP kinase Sty1 pathways control the onset of autophagy. Furthermore, we identify an unexpected phenotype of autophagy-defective mutants, namely their inability to survive in the absence of leucine when biosynthesis of this amino acid is impaired.
Reference: Corral-Ramos C, Barrios R, Ayté J, Hidalgo E (2022) TOR and MAP kinase pathways synergistically regulate autophagy in response to nutrient depletion in fission yeast. Autophagy 18:375-390.
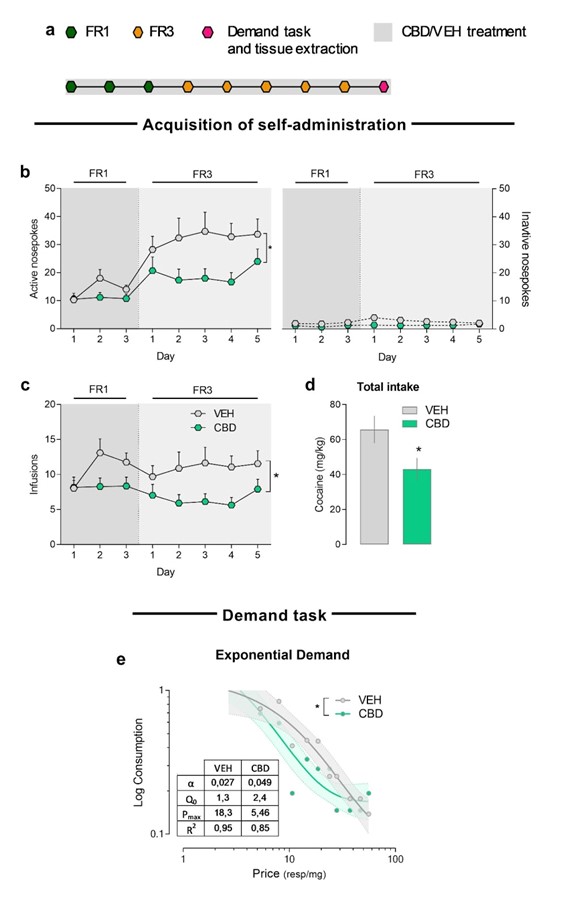
Cocaine is a highly consumed drug worldwide which directly targets brain areas involved in reinforcement processing and motivation. Cannabidiol is a phytocannabinoid that exerts protecting effects upon cocaine-induced addictive behavior, although many questions about the mechanisms of action and the specific affected processes remain unknown. Moreover, its effects on cue-induced cocaine-craving incubation have never been addressed. The present study aimed to assess the effects of cannabidiol (20 mg/kg, i.p.) administered during the acquisition of cocaine self-administration (0.75 mg/kg/infusion) and demand task or during cocaine abstinence and craving. Moreover, we measured the alterations in expression of AMPAR subunits and ERK1/2 phosphorylation due to cannabidiol treatment or cocaine withdrawal. Our results showed that cannabidiol reduced cocaine intake when administered during the acquisition phase of the self-administration paradigm, increased behavioral elasticity and reduced motivation for cocaine in a demand task. Cannabidiol also reduced GluA1/2 ratio and increased ERK1/2 phosphorylation in amygdala. No effects over cocaine-craving incubation were found when cannabidiol was administered during abstinence. Furthermore, cocaine withdrawal induced changes in GluA1 and GluA2 protein levels in the prelimbic cortex, ventral striatum and amygdala, as well as a decrease in ERK1/2 phosphorylation in ventral striatum. Taken together, our results show that cannabidiol exerts beneficial effects attenuating the acquisition of cocaine self-administration, in which an operant learning process is required. However, cannabidiol does not affect cocaine abstinence and craving.
Reference: Alegre-Zurano L, Berbegal-Sáez P, Luján MÁ, Cantacorps L, Martín-Sánchez A, García-Baos A, Valverde O (2022) Cannabidiol decreases motivation for cocaine in a behavioral economics paradigm but does not prevent incubation of craving in mice. Biomed Pharmacother.;148:112708.

Citizen consultations are public participation mechanisms designed to inform public policy and promote public dialogue. This article describes a deliberative consultation conducted within the CONCISE project framework. The aim was to gather qualitative knowledge about the means and channels through which European citizens acquire science-related knowledge, and how these influence their opinions and perceptions with respect to four socially relevant topics: vaccines, complementary and alternative medicine, genetically modified organisms, and climate change. In 2019, the CONCISE project carried out five citizen consultations in Poland, Slovakia, Spain, Italy and Portugal to explore the understanding of nearly 500 citizens, enabling the development of a standard for the carrying out of citizen consultations on science communication.
Reference: Llorente C, Revuelta G, Dziminska M, Warwas I, Krzewińska A, Moreno C (2022) A standard for public consultation on science communication: the CONCISE project experience. JCOM 21(03), N02.
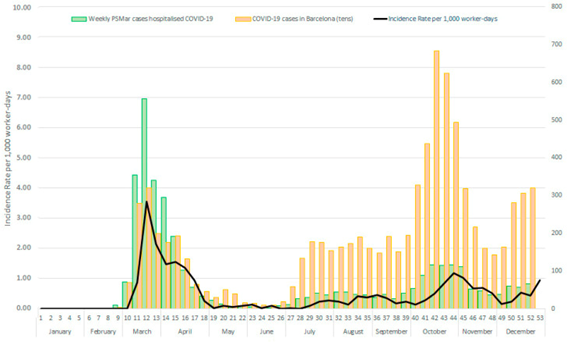
Figure: COVID-19 IR per 1000 worker-days among PSMar healthcare workers, hospitalised COVID-19 cases, and COVID-19 cases in Barcelona city by weeks.
Healthcare workers have been and still are at the forefront of COVID-19 patient care. Their infection had direct implications and caused important challenges for healthcare performance. The aim of this study is to assess the impact of non-pharmacological preventive measures against COVID-19 among healthcare workers. This study is based on a dynamic cohort of healthcare workers (n = 5543) who had been hired by a Spanish hospital for at least one week during 2020. Negative binomial regression models were used to estimate the incidence rate and the rate ratio (RR) between the two waves (defined from 15 March to 21 June and from 22 June to 31 December), considering natural immunity during the first wave and contextual variables. All models were stratified by socio-occupational variables. The average COVID-19 incidence rate per 1000 worker-days showed a significant reduction between the two waves, dropping from 0.82 (CI95%: 0.73–0.91) to 0.39 (0.35–0.44). The adjusted RR was 0.54 (0.48–0.87) when natural immunity was acquired during the first wave, and contextual variables were considered. The significant reduction of the COVID-19 incidence rate could be explained mainly by improvement in the non-pharmacological preventive interventions. It is needed to identify which measures were more effective. Young workers and those with a replacement contract were identified as vulnerable groups that need greater preventive efforts. Future preparedness plans would benefit from these results.
Reference: Utzet M, Benavides FG, Villar R, Burón A, Sala M, López LE, Gomar P, Castells X, Diaz P, Ramada JM, Serra C (2022) Non-Pharmacological Preventive Measures Had an Impact on COVID-19 in Healthcare Workers before the Vaccination Effect: A Cohort Study. Int J Environ Res Public Health 19(6):3628.
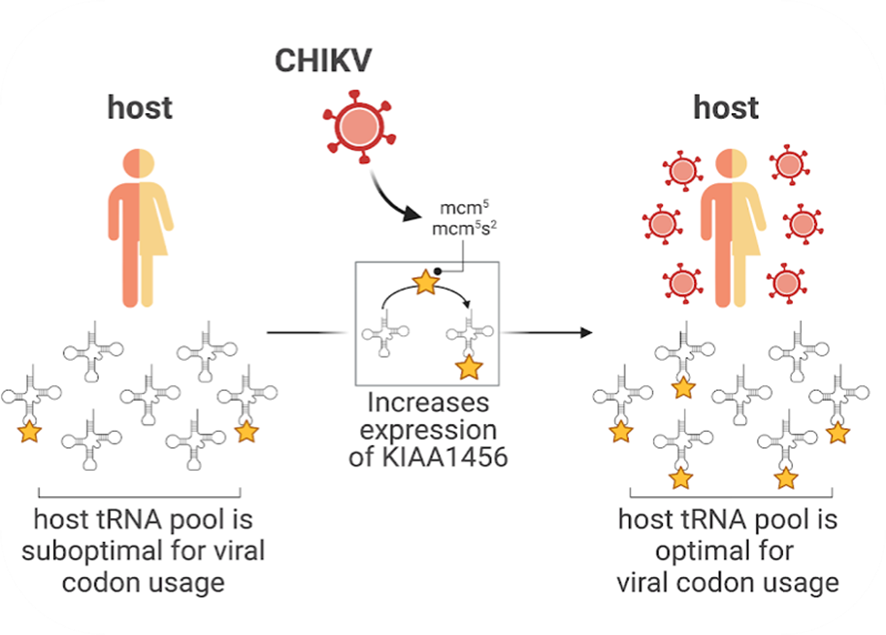
Ample evidence indicates that codon usage bias regulates gene expression. How viruses, such as the emerging mosquito-borne Chikungunya virus (CHIKV), express their genomes at high levels despite an enrichment in rare codons remains a puzzling question. Using ribosome footprinting, we analyze translational changes that occur upon CHIKV infection. We show that CHIKV infection induces codon-specific reprogramming of the host translation machinery to favor the translation of viral RNA genomes over host mRNAs with an otherwise optimal codon usage. This reprogramming was mostly apparent at the endoplasmic reticulum, where CHIKV RNAs show high ribosome occupancy. Mechanistically, it involves CHIKV-induced overexpression of KIAA1456, an enzyme that modifies the wobble U34 position in the anticodon of tRNAs, which is required for proper decoding of codons that are highly enriched in CHIKV RNAs. Our findings demonstrate an unprecedented interplay of viruses with the host tRNA epitranscriptome to adapt the host translation machinery to viral production.
Reference: Jungfleisch J, Böttcher R, Talló-Parra M,Pérez-Vilaró G, Merits A, Novoa EM , Díez J (2022) CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nature Communications, 13(1):4725.
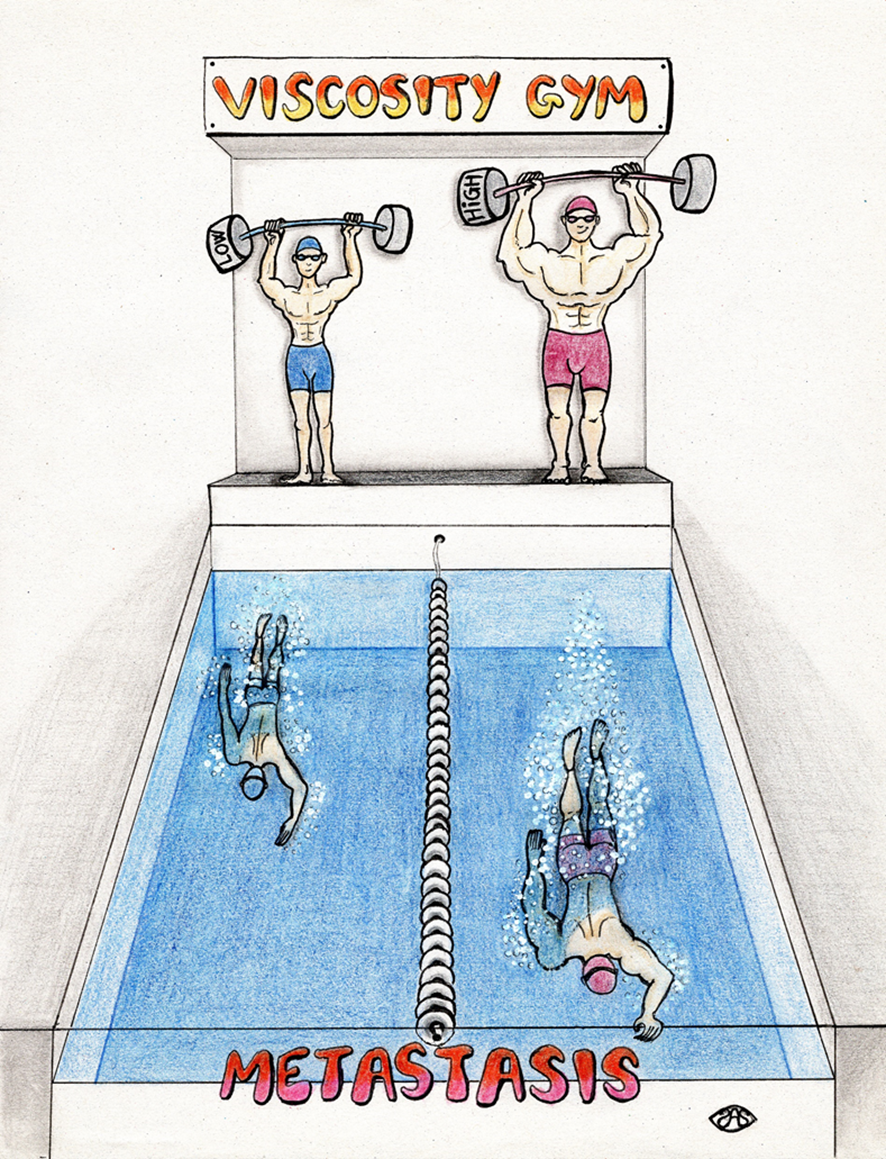
Cells respond to physical stimuli, such as stiffness, fluid shear stress and hydraulic pressure. Extracellular fluid viscosity is a key physical cue that varies under physiological and pathological conditions, such as cancer. However, its influence on cancer biology and the mechanism by which cells sense and respond to changes in viscosity are unknown. Here we demonstrate that elevated viscosity counterintuitively increases the motility of various cell types on two-dimensional surfaces and in confinement, and increases cell dissemination from three-dimensional tumour spheroids. Increased mechanical loading imposed by elevated viscosity induces an actin-related protein 2/3 (ARP2/3)-complex-dependent dense actin network, which enhances Na+/H+ exchanger 1 (NHE1) polarization through its actin-binding partner ezrin. NHE1 promotes cell swelling and increased membrane tension, which, in turn, activates transient receptor potential cation vanilloid 4 (TRPV4) and mediates calcium influx, leading to increased RHOA-dependent cell contractility. The coordinated action of actin remodelling/dynamics, NHE1-mediated swelling and RHOA-based contractility facilitates enhanced motility at elevated viscosities. Breast cancer cells pre-exposed to elevated viscosity acquire TRPV4-dependent mechanical memory through transcriptional control of the Hippo pathway, leading to increased migration in zebrafish, extravasation in chick embryos and lung colonization in mice. Cumulatively, extracellular viscosity is a physical cue that regulates both short- and long-term cellular processes with pathophysiological relevance to cancer biology.
Reference: Bera K, Kiepas A, Godet I, Li Y, Mehta P, Ifemembi B, Paul CD, Sen A, Serra SA, Stoletov K, Tao J, Shatkin G, Lee SJ, Zhang Y, Boen A, Mistriotis P, Gilkes DM, Lewis JD, Fan CM, Feinberg AP, Valverde MA, Sun SX, Konstantopoulos K (2022) Extracellular fluid viscosity enhances cell migration and cancer dissemination. Nature 611, 365–373
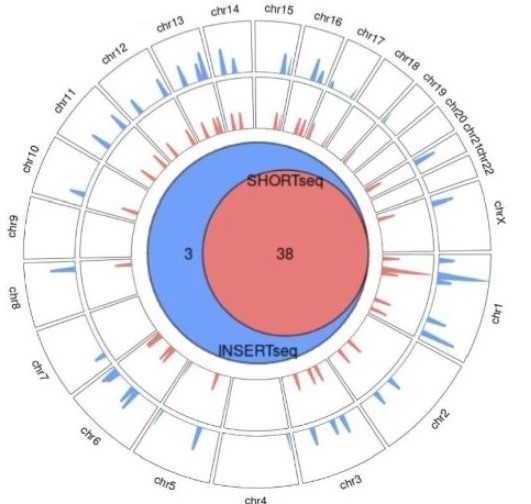
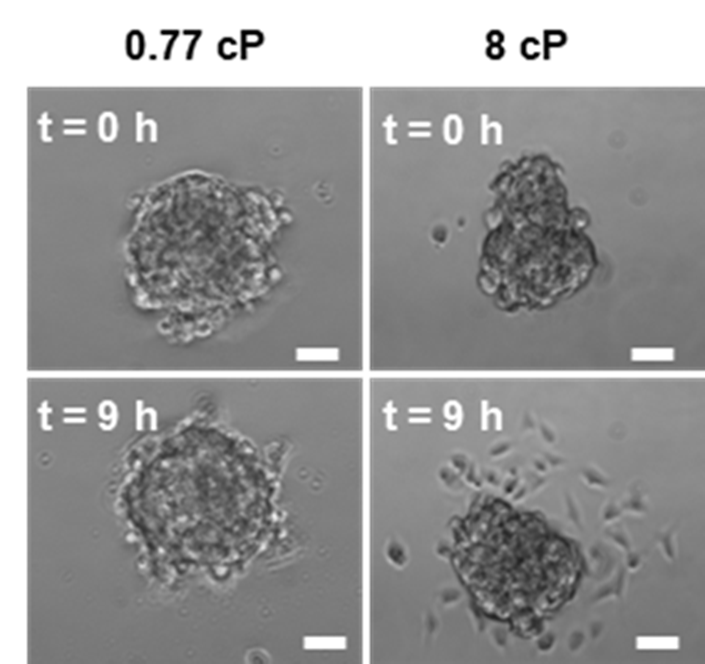
Comprehensive characterisation of genome engineering technologies is relevant for their development and safe use in human gene therapy. Short-read based methods can overlook insertion events in repetitive regions. We develop INSERT-seq, a method that combines targeted amplification of integrated DNA, UMI-based correction of PCR bias and Oxford Nanopore long-read sequencing for robust analysis of DNA integration. The experimental pipeline improves the number of mappable insertions at repetitive regions by 4.8–7.3% and larger repeats are processed with a computational peak calling pipeline. INSERT-seq is a simple, cheap and robust method to quantitatively characterise DNA integration in diverse ex vivo and in vivo samples.
Reference: Ivančić D, Mir-Pedrol J, Jaraba-Wallace J, Rafel N, Sanchez-Mejias A, Güell M (2022) INSERT-seq enables high-resolution mapping of genomically integrated DNA using Nanopore sequencing. Genome Biol 23, 227
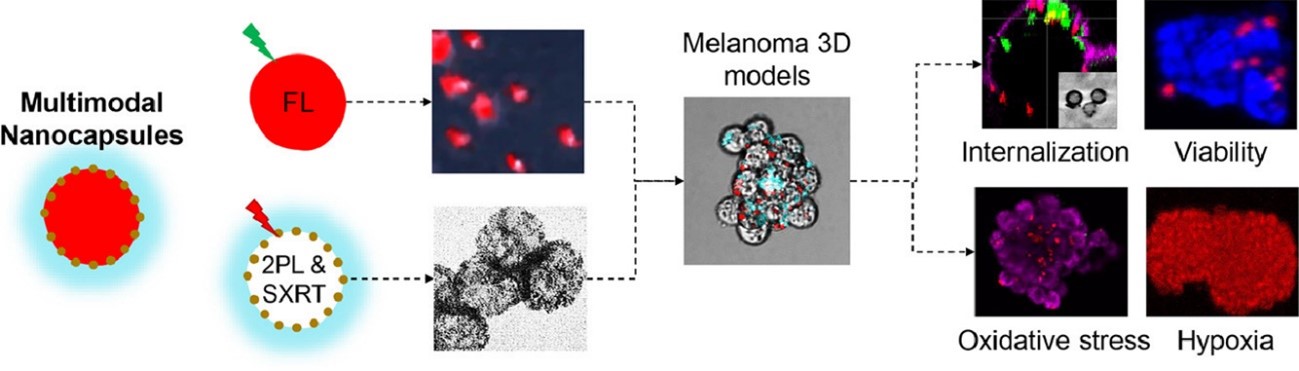
We report the synthesis of plasmonic nanocapsules and the cellular responses they induce in 3D melanoma models for their perspective use as a photothermal therapeutic agent. The wall of the nanocapsules is composed of polyelectrolytes. The inner part is functionalized with discrete gold nanoislands. The cavity of the nanocapsules contains a fluorescent payload to show their ability for loading a cargo. The nanocapsules exhibit simultaneous two-photon luminescent, fluorescent properties and X-ray contrasting ability. The average fluorescence lifetime (τ) of the nanocapsules measured with FLIM (0.3 ns) is maintained regardless of the intracellular environment, thus proving their abilities for bioimaging of models such as 3D spheroids with a complex architecture. Their multimodal imaging properties are exploited for the first time to study tumorspheres cellular responses exposed to the nanocapsules. Specifically, we studied cellular uptake, toxicity, intracellular fate, generation of reactive oxygen species, and effect on the levels of hypoxia by using multi-photon and confocal laser scanning microscopy. Because of the high X-ray attenuation and atomic number of the gold nanostructure, we imaged the nanocapsule-cell interactions without processing the sample. We confirmed maintenance of the nanocapsules’ geometry in the intracellular milieu with no impairment of the cellular ultrastructure. Furthermore, we observed the lack of cellular toxicity and no alteration in oxygen or reactive oxygen species levels. These results in 3D melanoma models contribute to the development of these nanocapsules for their exploitation in future applications as agents for imaging-guided photothermal therapy.
Reference: Zamora-Perez P, Xiao C, Sanles-Sobrido M, Rovira-Esteva M, Conesa JJ, Mulens-Arias V, Jaque D, Rivera-Gil P (2022) Multiphoton imaging of melanoma 3D models with plasmonic nanocapsules. Acta Biomaterialia, 142, 308-19.
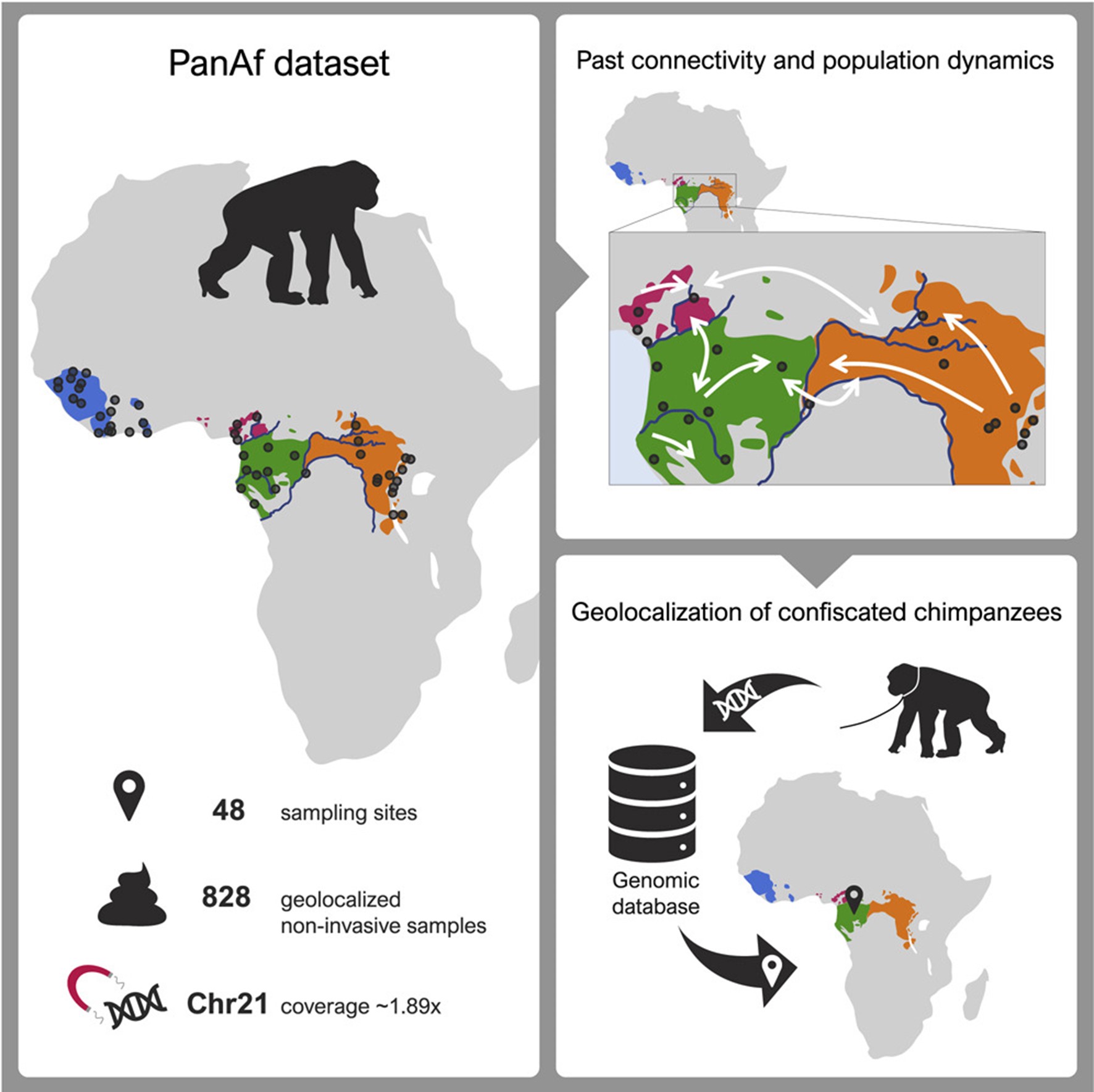
Knowledge on the population history of endangered species is critical for conservation, but whole-genome data on chimpanzees (Pan troglodytes) is geographically sparse. Here, we produced the first non-invasive geolocalized catalog of genomic diversity by capturing chromosome 21 from 828 non-invasive samples collected at 48 sampling sites across Africa. The four recognized subspecies show clear genetic differentiation correlating with known barriers, while previously undescribed genetic exchange suggests that these have been permeable on a local scale. We obtained a detailed reconstruction of population stratification and fine-scale patterns of isolation, migration, and connectivity, including a comprehensive picture of admixture with bonobos (Pan paniscus). Unlike humans, chimpanzees did not experience extended episodes of long-distance migrations, which might have limited cultural transmission. Finally, based on local rare variation, we implement a fine-grained geolocalization approach demonstrating improved precision in determining the origin of confiscated chimpanzees.
Reference: Fontsere C, Kuhlwilm M, Morcillo-Suarez C, et al., Marques-Bonet T (2022) Population dynamics and genetic connectivity in recent chimpanzee history. Cell Genom (6):100133.

Peru hosts extremely diverse ecosystems which can be broadly classified into the following three major ecoregions: the Pacific desert coast, the Andean highlands, and the Amazon rainforest. Since its initial peopling approximately 12,000 years ago, the populations inhabiting such ecoregions might have differentially adapted to their contrasting environmental pressures. Previous studies have described several candidate genes underlying adaptation to hypobaric hypoxia among Andean highlanders. However, the adaptive genetic diversity of coastal and rainforest populations has been less studied. Here, we gathered genome-wide single-nucleotide polymorphism-array data from 286 Peruvians living across the three ecoregions and analyzed signals of recent positive selection through population differentiation and haplotype-based selection scans. Among highland populations, we identify candidate genes related to cardiovascular function (TLL1, DUSP27, TBX5, PLXNA4, SGCD), to the Hypoxia-Inducible Factor pathway (TGFA, APIP), to skin pigmentation (MITF), as well as to glucose (GLIS3) and glycogen metabolism (PPP1R3C, GANC). In contrast, most signatures of adaptation in coastal and rainforest populations comprise candidate genes related to the immune system (including SIGLEC8, TRIM21, CD44, and ICAM1 in the coast; CBLB and PRDM1 in the rainforest; and BRD2, HLA-DOA, HLA-DPA1 regions in both), possibly as a result of strong pathogen-driven selection. This study identifies candidate genes related to human adaptation to the diverse environments of South America.
Reference: Caro-Consuegra R, Nieves-Colón MA, Rawls E, Rubin-de-Celis V, Lizárraga B, Vidaurre T, Sandoval K, Fejerman L, Stone AC, Moreno-Estrada A, Bosch E (2022) Uncovering Signals of Positive Selection in Peruvian Populations from Three Ecological Regions. Molecular Biology and Evolution, Volume 39, Issue 8, August 2022, msac158.
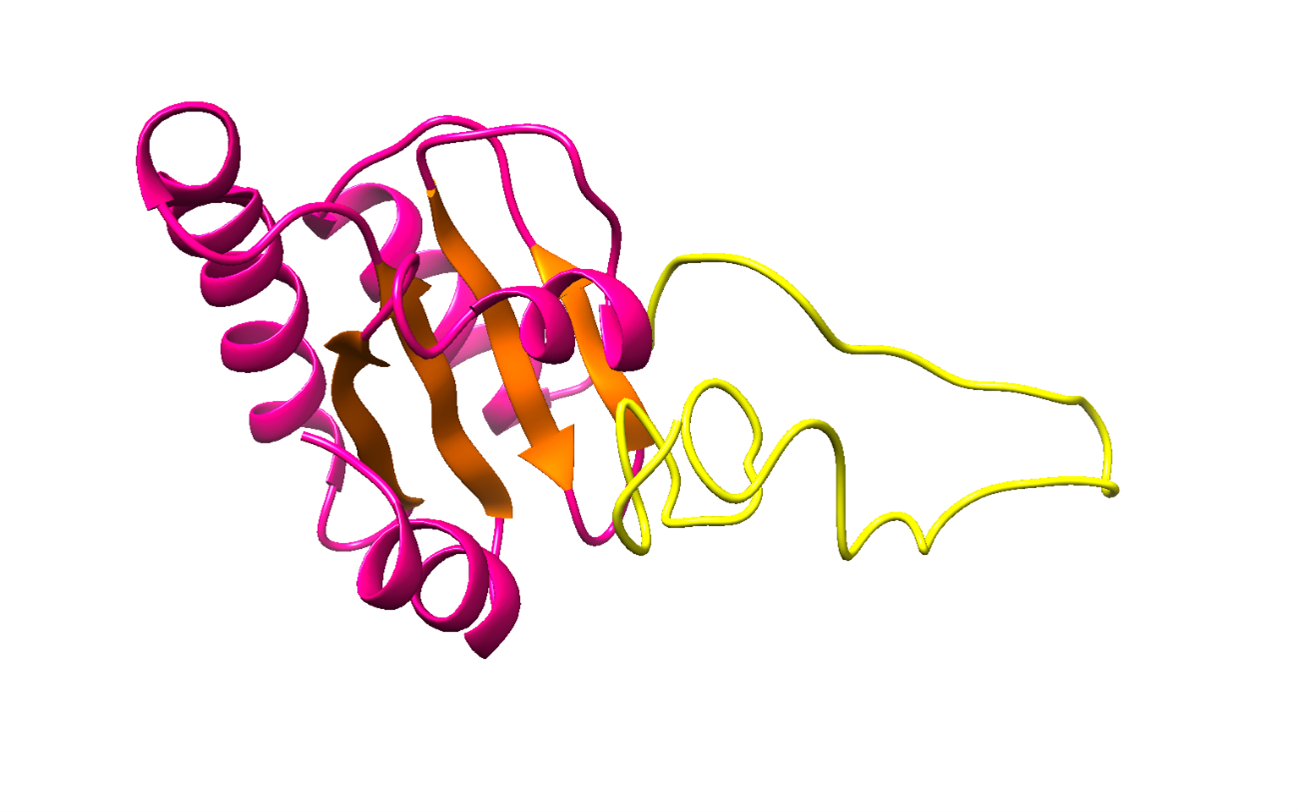
RNA-binding proteins (RBPs) have been relatively overlooked in cancer research despite their contribution to virtually every cancer hallmark. Here, we use RNA interactome capture (RIC) to characterize the melanoma RBPome and uncover novel RBPs involved in melanoma progression. Comparison of RIC profiles of a non-tumoral versus a metastatic cell line revealed prevalent changes in RNA-binding capacities that were not associated with changes in RBP levels. Extensive functional validation of a selected group of 24 RBPs using five different in vitro assays unveiled unanticipated roles of RBPs in melanoma malignancy. As proof-of-principle we focused on PDIA6, an ER-lumen chaperone that displayed a novel RNA-binding activity. We show that PDIA6 is involved in metastatic progression, map its RNA-binding domain, and find that RNA binding is required for PDIA6 tumorigenic properties. These results exemplify how RIC technologies can be harnessed to uncover novel vulnerabilities of cancer cells.
Reference: Mestre-Farràs N, Guerrero S, Bley N, Rivero E, Coll O, Borràs E, Sabido E, Indacochea A, Casillas-Serra C, Järvelin AI, Oliva B, Castello A, Hüttelmaier S, Gebauer F (2022) Melanoma RBPome identification reveals PDIA6 as an unconventional RNA-binding protein involved in metastasis. Nucleic acids research; 50, 14, 8207-8225.
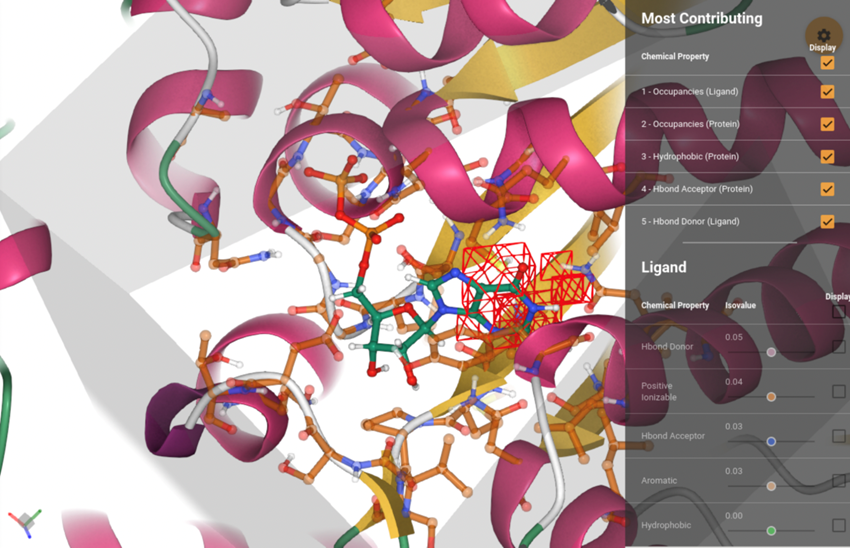
The protein−ligand complex is displayed, with the attributions of the most contributing voxels superimposed. The attributions for the different channels can be seen individually using the corresponding sliders in the menu on the right, which display isosurfaces at different isovalues. The full protein is shown in a cartoon representation, while residues in the binding site (defined by being within 5 Å to the ligand) are shown in a transparent ball−stick representation (only heavy atoms and polar hydrogens). The all-atom representation of the ligand is shown in a bold ball−stick. The region of space seen by the model (voxelization cube) is delimited by a transparent, gray box.
RNA-binding proteins (RBPs) hDeep learning has been successfully applied to structure-based protein–ligand affinity prediction, yet the black box nature of these models raises some questions. In a previous study, we presented KDEEP, a convolutional neural network that predicted the binding affinity of a given protein–ligand complex while reaching state-of-the-art performance. However, it was unclear what this model was learning. In this work, we present a new application to visualize the contribution of each input atom to the prediction made by the convolutional neural network, aiding in the interpretability of such predictions. The results suggest that KDEEP is able to learn meaningful chemistry signals from the data, but it has also exposed the inaccuracies of the current model, serving as a guideline for further optimization of our prediction tools.
Reference: Varela-Rial A, Maryanow I, Majewski M, Doerr S, Schapin N, Jiménez-Luna J, De Fabritiis G (2022) PlayMolecule Glimpse: Understanding Protein-Ligand Property Predictions with Interpretable Neural Networks. J Chem Inf Model; 62(2): 225-231
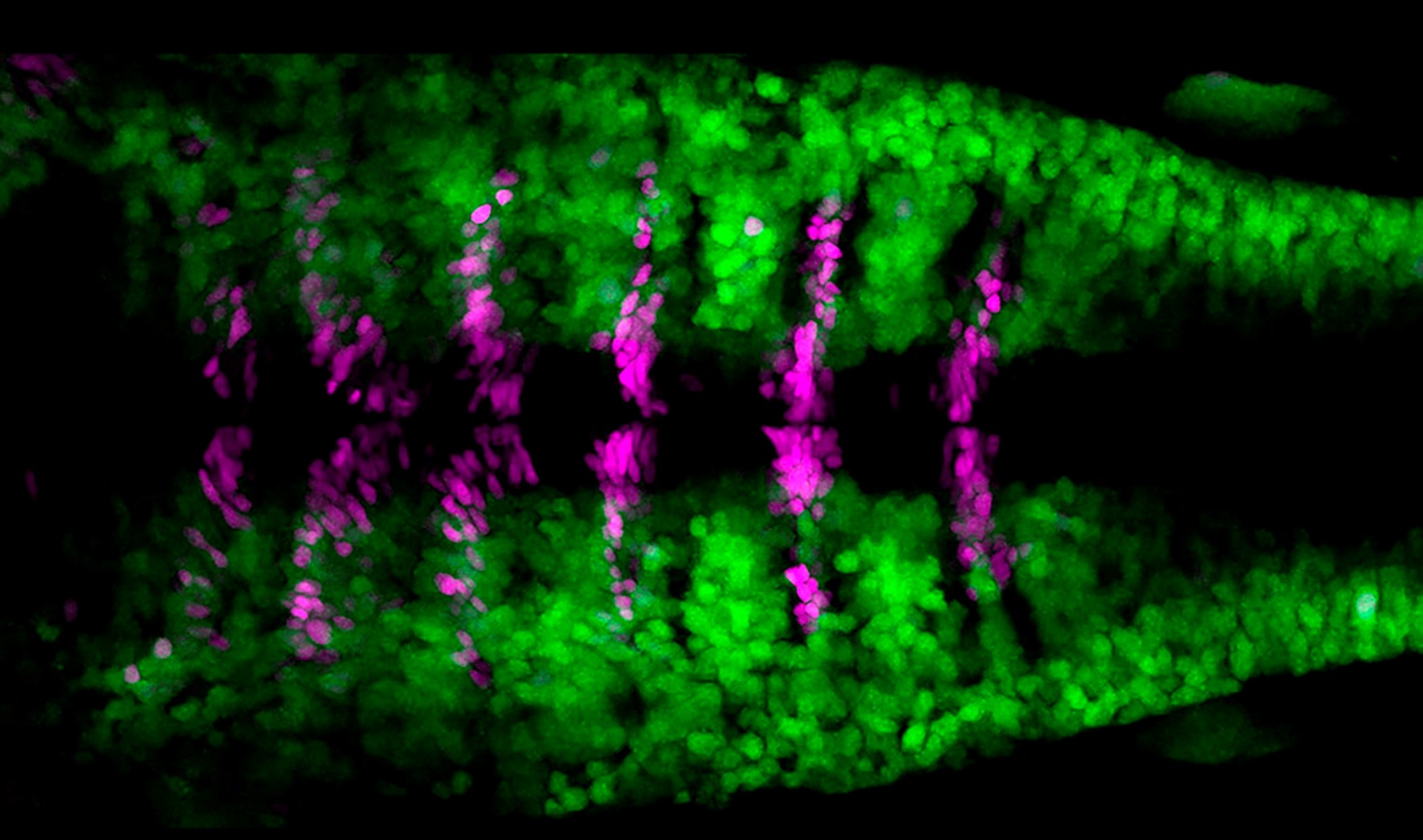
Elucidating the cellular and molecular mechanisms that regulate the balance between progenitor cell proliferation and neuronal differentiation in the construction of the embryonic brain demands the combination of cell lineage and functional approaches. Here, we generate the comprehensive lineage of hindbrain boundary cells by using a CRISPR-based knockin zebrafish transgenic line that specifically labels the boundaries. We unveil that boundary cells asynchronously engage in neurogenesis undergoing a functional transition from neuroepithelial progenitors to radial glia cells, coinciding with the onset of Notch3 signaling that triggers their asymmetrical cell division. Upon notch3 loss of function, boundary cells lose radial glia properties and symmetrically divide undergoing neuronal differentiation. Finally, we show that the fate of boundary cells is to become neurons, the subtype of which relies on their axial position, suggesting that boundary cells contribute to refine the number and proportion of the distinct neuronal populations.
Reference: Hevia CF, Engel-Pizcueta C, Udina F, Pujades C (2022) The neurogenic fate of the hindbrain boundaries relies on Notch3-dependent asymmetric cell divisions. Cell Rep 39(10):110915.
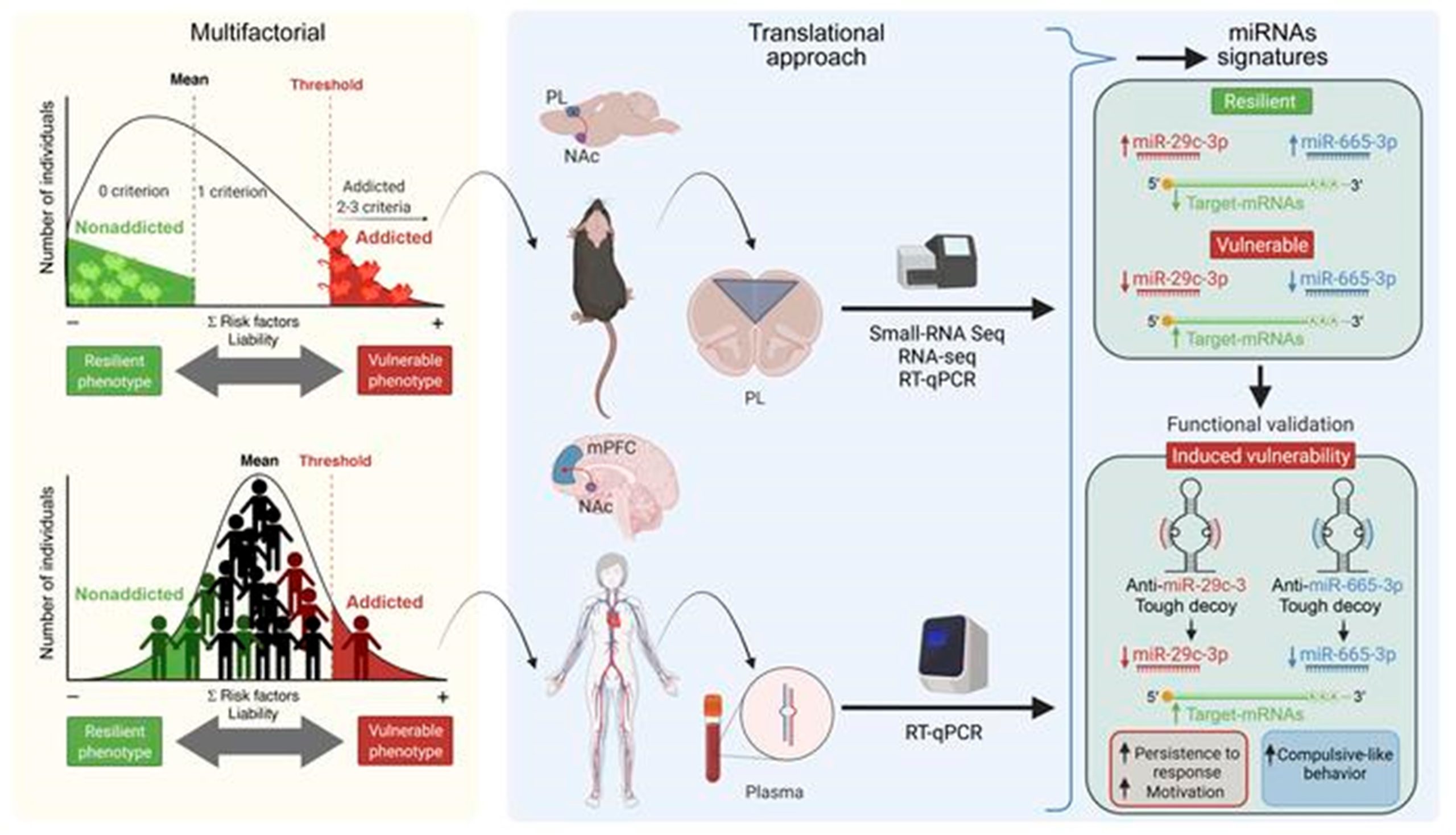
Food addiction is characterized by a loss of behavioral control over food intake and is associated with obesity and other eating disorders. The mechanisms underlying this behavioral disorder are largely unknown. We aimed to investigate the changes in miRNA expression promoted by food addiction in animals and humans and their involvement in the mechanisms underlying the behavioral hallmarks of this disorder. We found sharp similitudes between miRNA signatures in the medial prefrontal cortex (mPFC) of our animal cohort and circulating miRNA levels in our human cohort, which allowed us to identify several miRNAs of potential interest in the development of this disorder. Tough decoy (TuD) inhibition of miRNA-29c-3p in the mouse mPFC promoted persistence of the response and enhanced vulnerability to developing food addiction, whereas miRNA-665-3p inhibition promoted compulsion-like behavior and also enhanced food addiction vulnerability. In contrast, we found that miRNA-137-3p inhibition in the mPFC did not lead to the development of food addiction. Therefore, miRNA-29c-3p and miRNA-665-3p could be acting as protective factors with regard to food addiction. We believe the elucidation of these epigenetic mechanisms will lead to advances toward identifying innovative biomarkers and possible future interventions for food addiction and related disorders based on the strategies now available to modify miRNA activity and expression.
Reference: García-Blanco A, Domingo-Rodriguez L, Cabana-Domínguez J, Fernández-Castillo N, Pineda-Cirera L, Mayneris-Perxachs J, Burokas A, Espinosa-Carrasco J, Arboleya S, Latorre J, Stanton C, Cormand B, Fernández-Real JM, Martín-García E, Maldonado R (2022) miRNA signatures associated with vulnerability to food addiction in mice and humans. J Clin Invest. 132(10):e156281.